| المجلد 8 ,
العددان 1 و 2
, رمضان 1437 - تموز (يوليو) 2016 |
| |
| التحري عن جينmecA وجينvanA في العنقودية الذهبية والعنقودية الجلدية من العينات المرضية |
| Detection of mecA Gene, vanA Gene in Staphylococcus Aureus and Staphylococcus Epidermidis from Clinical Samples |
| د. مروان حسين، و د. عدنان نظام، و د. سمير أبو اصبع |
| Hussain M; Nizam A. and Abou-Isba S. |
قسم علم الحياة النباتية- كلية العلوم- جامعة دمشق- سورية
Department of plant Biology- Faculty of Science- Damascus University
|
| الملخص Abstract |
| هدفت الدراسة الحالية إلى استعراف سلالات كل من العنقودية الذهبية والعنقودية الجلدية، واستقصاء جين المقاومة للميتيسلين mecA والفانكوميسين vanA في هذه الجراثيم، واكتشاف مقاومتها للمضادات الحيوية. جرى الحصول على العينات من ثلاث مستشفيات: المواساة، الأطفال، والأمراض الجلدية والزهرية في الفترة ما بين آب 2013 وحتى حزيران 2014. ولقد اكتشفت 46 سلالة من العنقودية الذهبية والجلدية، حيث أظهرت نتائج PCR عصابات مفردة لكل من جين nuc، وجين mecA، وجين vanA، التي ترمز مناطق طولها تقريباً 270 bp، و533 bp، و bp1100، على الترتيب. وكانت نسبة وجود جين mecA في سلالات العنقودية الذهبية (MRSA) 15%، ونسبة وجود جين vanA كانت 2.5%. وكانت نسب المقاومة للمضادات الحيوية كالآتي: الأوغمانتين AMC (65%)، أمبيسيلين (70%)، أموكساسيلين (55%)، جنتاميسين (30%)، سيفترياكسون (62.5%)، بيفلوكساسين (7.5%)، أوفلوكساسين (7.5%)، فانكوميسين (2.5%). وكان التركيز الأصغري المثبط للميتيسلينMIC بالنسبة للعنقودية الذهبية ضمن مجال بين 16-32 مكغ/مل، ما عدا السلالات المقاومة فكانت أكثر من 256 مكغ/مل. بينما MIC للفانكوميسين كان ضمن مجال بين 4-8 مكغ/مل، ما عدا السلالات المقاومة فكانت أكثر من 256 مكغ/مل. أما التركيز الأصغري المثبط للميتيسلين بالنسبة لكل سلالات العنقودية الجلدية فكان ضمن مجال بين 8-16 مكغ/مل. |
| Current study aimed to identify S. aureus and S. epidermidis strains, and investigate of methicillin resistant gene mecA and vancomycin vanA gene in these bacteria and detection the antimicrobial resistance. The samples were obtained from three hospitals: Al-Mouwasat, Children, and Dermal Venereal Diseases between August 2013 through June 2014. Forty six strains of S. aureus and S. epidermidis were detected, PCR product appeared a single DNA band for nuc, mecA and vanA genes, which coded areas equal to nearly 270 bp, 533 bp, and 1100 bp respectively. The percentage of presence of mecA gene in S. aureus strains (MRSA) was 15% and vanA gene (VRSA) was 2.5%. The percentages of antibiotic resistance were: AMC (65%), AM (70%), AX (55%), GN (30%), CRO (62.5%), PE (7.5%), OFX (7.5%) and VA (2.5%). The MIC of S. aureus was ranging between 16 – 32 μg ml-1, except resistant strains was >256 μg ml-1 while MIC for vancomycin was ranging between 4 – 8 μg ml-1, except resistant strains was >256 μg ml-1. Whereas the MIC for methicillin of all strains of S. epidermidis was ranging between 8 – 16 μg ml-1. |
| Introduction |
Staphylococcus aureus is a major pathogen for humans, and it is a frequent pathogen in nosocomial infections and limited outbreaks in hospitals. Almost every person will have some type of S. aureus infection during a lifetime, ranging in severity from food poisoning or minor skin infections to severe life-threatening infections (1, 2). S. aureus is a zoonotic and human pathogen that can cause a wide range of human infections from pimples and boils to pneumonia, food poisoning, bloodstream, respiratory tract, UTIs and surgical wound infections. Forthmore, skin infections, osteomyelitis and acute endocarditis (3, 4).
Coagulase Negative Staphylococci (CNS), especially Staphylococcus epidermidis cause mainly foreign body infections like intravasal catheters, continuous ambulant peritoneal dialysis (CAPD) catheters, endoprostheses, metal plates and screws in osteosynthesis, cardiac pacemakers, artificial heart valves, and shunt valves, in addition, S epidermidis is more often resistant to antimicrobial drugs in these cases (3, 5), where Palazzo et al. (6) published the first report of vancomycin-resistant Staphylococci isolated from healthy carriers; Wolk et al. (7) showed rapid detection and sensitivity of S. aureus and MRSA in wound and blood cultures; Al-Obeid et al. (8) showed the first detection of an invasive S. aureus strain with reduced susceptibility to glycopeptides; Bhargava and Zhang (9) isolated and characterized 87 strains of CNS from food animals by molecular methods and studied antimicrobial susceptibility; Gutierrez et al. (10) identified the hospitalized children with Staphylococcal infections through the California office of statewide health planning and development discharge database between 1985 – 2009; De Matos et al. (11) evaluated the synergistic activity of antimicrobial drugs against lineages of forty two MRSA isolates carrying SCCmec IV. The aim of current study was (i) identification of S. aureus and S. epidermidis strains, then investigation of mecA and vanA genes in these bacteria. (ii) detection the antimicrobial resistance by disc diffusion with MICs in these bacteria.
|
| Materials and Methods
Bacterial strains
|
Between 8/2013 – 6/2014 a total of 46 staphylococcal strains were isolated from clinical samples (ear, urine, eye, CSF, brain peritoneal shunt, bronchial lavages, pleural effusion , abscess, furuncle, bullous, Pus, Wound, face and hand) from Damascus city. The samples were obtained from three hospitals: Al-Mouwasat, Children, and
Dermal and Venereal Diseases.
Identification of strains
The isolates obtained from the clinical specimens were seeded onto Nutrient Agar NA(Abtec, England) and stained by the Gram stain. According to the Bergey’s manual (12), the genus Staphylococcus was differentiated from Streptococcus by testing catalase test. S. aureus was distinguished from other species, by seeded onto mannitol salt agar MSA (Biolife, Italy), blood agar BA (Abtec, England), coagulase slide test (Sigma, Germany) according to the manufacturer’s instructions. Further biochemical identification of Staphylococci to the species level was performed using API Staph (BioMérieux, France). The plates were incubated at 37ºC for 24 h. (12, 13).
After species confirmation, the isolates were stored on NA slants at 4ºC and in nutrient broth NB (SRL, India) with 20% glycerol (v/v) at -30ºC freezer. Routinely, fresh bacterial cultures were obtained from frozen stocks before each experiment and were grown at 24ºC in NA (14).
|
| Molecular methods
DNA extraction
|
Staphylococci were grown in 50 ml Nutreint Broth NB (SRL, India) for 24 h., then DNA extraction was done according to )15, 16) with simple modifications.
PCR conditions for Detection of nuc, mecA, and vanA genes
PCR was performed using S. aureus species-specific primers (Alpha DNA, Canada) for the nuc gene, which codes for thermostable nuclease using the primers, F(5`-GCGATTGATGGTGAT ACGGTT-3`) and R (5`-AGCCAAGC CTTGACGAACTAAAGC-3`) as described by (17), generating ~ 270 bp. PCR amplification of mecA gene was performed on 0.5 μg of genomic DNA, primers for the detection of mecA were F (5`-AGTTGTAGTTGTCGGGTTT-3`) and R(5`-AGTGGAACGAAGGTATC ATC-3`) (17) generating a 533 bp. Finally the vanA gene was amplified with specific primers. The primers for amplification of vanA were F (5`- ATG AATAGAATAAAAGTTGC-3`) and R (5`-TCACCCCTTTAACGCTAATA-3`) as described by (18) generating a 1100 bp. The PCR was carried out in 25μl volumes under the following conditions: 100 ng genomic DNA, 12.5 μl of One PCRTM Master Mix (which contains Taq DNA polymerase, PCR buffer, dNTPs, gel loading dyes and fluorescence dye), 250 nM of each primer, and nuclease free water was added to make up the final volume of 25 μl. The reaction conditions consisted of one initial denaturation cycle at 94ºC for 3 min., 45 cycles of 94ºC for 1 min., 55ºC (for nuc and vanA) and 58 ºC (for mecA) for 1 min and 72ºC for 2 min., and a final extension step at 72ºC for 10 min. then cold 4ºC. Amplifications were performed in Bioer thermal cycler (China). The amplified products were run on a 1.25% agarose gel and visualized on a u.v. transilluminator.
Antibiotic susceptibility
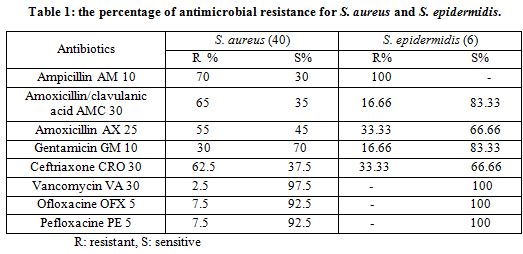
All tested Staphylococci samples were tested against susceptibility for eight antibiotics using Kirby-Bauer disc diffusion assay following the recommendations of the Clinical and Laboratory Standards Institute (19). Isolates were streaked on Mueller-Hinton agar (Merck, Germany) and incubated at 37˚C for 24 h. with the following antibiotic: amoxicillin-clavulanic acid (AMC, 30μg), (gentamicin, 10 μg), (ceftriaxone, 30 μg), (pefloxacine, 5 μg), (ofloxacine, 5 μg) and (ampicillin, 10 μg), (vancomycin, 30 μg,) (amoxicillin, 25µg). All discs were obtained from (Bio-analyze, Turkey). Inhibition zone diameters were measured and studied using norms of the National Committee on Clinical Laboratory Standards (NCCLS) (Table, 1).
MIC determination
Determination of MIC of methicillin and vancomycin (Sigma- Aldrich) were determined by broth dilution method using CLSI guidelines (19). The Antibiotics were dissolved in distilled water with 1 ml inoculum (0.5 McFarland) of Staphylococci and serial dilutions were made in NB ranging from 256 - 4 μg ml. All the tubes were incubated for 24 hrs. at 37ºC. MICs were determined as the lowest concentration of antibiotics that showed no visible growth of the Staphylococci.
|
| Results and Discussion
Bacterial Identification
|
All the strains S1 to S46 were found to be gram positive cocci in clusters and catalase test. All the strains of S. aureus were isolated from patients were Positive response for (mannitol fermentation, coagulase slide test). After confirmation the characteristics upon biochemical tests and API Staph. the results were submitted to 40 S. aureus. and 6 S. epidermidis.
Antibiotic susceptibility tests
Strains of S. aureus showed different level of resistance against some antibiotics. Inhibition zone diameters were measured depending on NCCLS. The results of antibiotic resistance tests of S. aureus was as follows: amoxicillin-clavulanic acid (65%), gentamicin (30%), ceftriaxone (62.5%), pefloxacine (7.5%), ofloxacine (7.5%) ampicillin (70%), vancomycin (2.5%) and amoxicillin (55%) (Table 1, Fig. 1, 2).
Hinton agar (Merck, Germany) and incubated at 37˚C for 24 h. with the following antibiotic: amoxicillin-clavulanic acid (AMC, 30μg), (gentamicin, 10 μg), (ceftriaxone, 30 μg), (pefloxacine, 5 μg), (ofloxacine, 5 μg) and (ampicillin, 10 μg), (vancomycin, 30 μg,) (amoxicillin, 25µg). All discs were obtained from (Bio-analyze, Turkey). Inhibition zone diameters were measured and studied using norms of the National Committee on Clinical Laboratory Standards (NCCLS) (Table, 1).
MIC determination
Determination of MIC of methicillin and vancomycin (Sigma- Aldrich) were determined by broth dilution method using CLSI guidelines (19). The Antibiotics were dissolved in distilled water with 1 ml inoculum (0.5 McFarland) of Staphylococci and serial dilutions were made in NB ranging from 256 - 4 μg ml. All the tubes were incubated for 24 hrs. at 37ºC. MICs were determined as the lowest between 8 – 16 μg ml-1. These results like some studies as a (6) which carried out in Brazil, where MIC was 4 μg ml for S. epidermidis, and study of (15), while different of study (17) where showed that some strains of S. aureus had a MIC >256 μg ml-1 and others had <8 μg ml-1 and against vancomycin was found that 53% of isolates had 4 μg ml-1.
|
 |
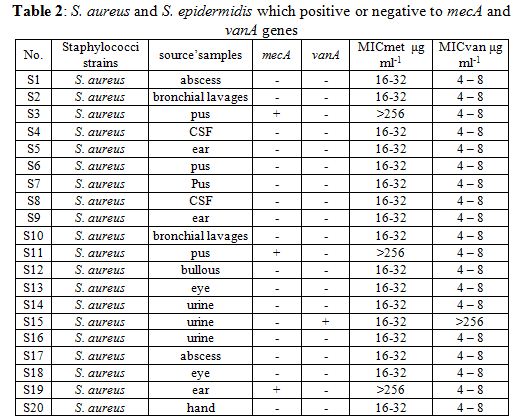
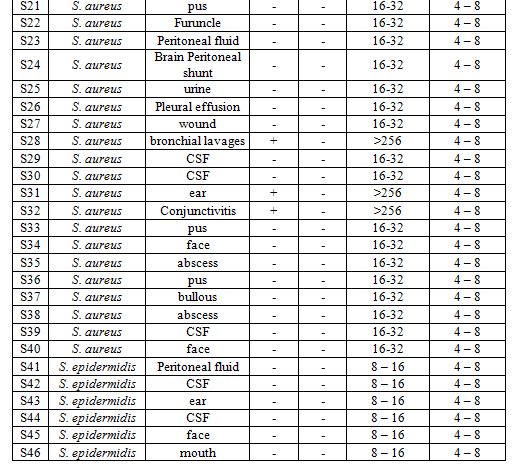
The MIC for methicillin of all strains of S. aureus was ranging between 16 – 32 μg ml-1 except the strains which carry mecA gene (S3, S11, S19, S28, S31, S32) was >256 μg ml-1, but MIC for vancomycin was ranging between 4 – 8 μg ml-1 except the strain which carry vanA gene (S15) was >256 μg ml-1. Whereas The MIC for methicillin of all strains of S. epidermidis was ranging between 8 – 16 μg ml-1. These results like some studies as a (6) which carried out in Brazil, where MIC was 4 μg ml for S. epidermidis, and study of (15), while different of study (17) where showed that some strains of S. aureus had a MIC >256 μg ml-1 and others had <8 μg ml-1 and against vancomycin was found that 53% of isolates had 4 μg ml-1.
Detection of nuc, mecA, and vanA genes
Genotyping have been used for the detection of clinical isolates to guide the management of nosocomial infections. However, a variety of markers can be used for typing and results accordingly reflect the choice of markers. In Current study it has been identified the S. aureus and S. epidermidis, then detected the mecA gene and vanA gene, moreover, the antimicrobial resistance by disc diffusion with MICs in these bacteria.
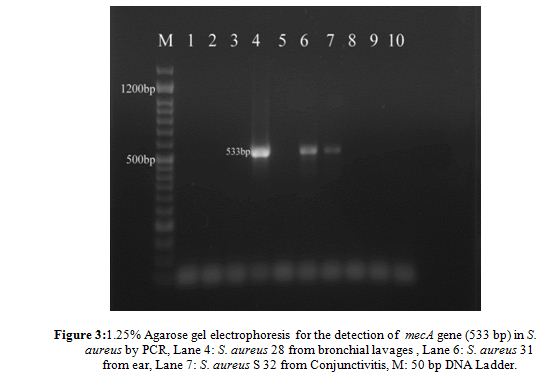
For molecular characterization the amplifying thermostable nuclease (nuc) gene encoded to enzyme thermonuclease and the PCR product appeared a single DNA band with a size equal to ~ 270 bp corresponding to the nuc gene. The mecA gene was detected in all strains by the PCR amplification method. PCR amplification of mecA gene was positive in 6 of 40 strains (MRSA): S3, S11, S19, S28, S31, S32, but the others 34 strains were MSSA Methicillin-sensitive Staph. aureus, where the percentage of presence of mecA was (15%) (Table 2, Fig. 3). While for S. epidermidis mecA gene was not founded in any strain. The strains which were carried out mecA gene showed resistance to amoxicillin, ampicillin, amoxicillin clavulanic acid.
By S. aureus and based on antibiotic resistance discs there are 28 strains resistant to ampicillin and 22 strains resistant to amoxicillin, but only 6 strains were carried out mecA gene, that is to say not all strains of S. aureus can be classified MRSA, like S1, S2, S6, S15 which were resistant to (AMC, AM, AX) but it didn’t carry mecA gene. The repression of mecA gene and the resulting absence of MR in some of the isolates could be due to several factors. Both genetic and environmental factors play a significant role in the expression of MR. While all strains of S. epidermidis were sensitive for all tested antibiotics and this agree with genetic results, where mecA gene didn’t found in any strains.
In contrast to previously reported MRSA isolates, Khan et al. (20) showed that 7 isolates of 17 were carried out by mecA gene; Buzaid et al. (21) reported that 31 % of S. aureus were MRSA in tertiary surgical and trauma hospital; Abd-El Hafez et al. (22) showed that 29 isolates of S. epidermidis were positive for the mecA gene, and these isolates showed several multidrug-resistance patterns; Bhargava and Zhang (9) detected the mecA gene in 60 of 87 CNS isolates and studied their antimicrobial susceptibility to some antibiotics; Lyer et al. (23) appeared that about 73 of 100 S. aureus were isolated from workers were positive for mecA gene; Sergelidis et al. (24) isolated methicillin-resistant staphylococcus spp. from ready to eat fish products and showed presence the mecA gene in two oxacillin-resistant strains and three (23.1%) of S. epidermidis.
PCR results of mecA gene gave sharp differentiation between many strains which help in determination of the suspected source of infection especially in nosocomial infection cases also in case of repeated infections with the same strain in case of treatment failure or insufficient disinfection.
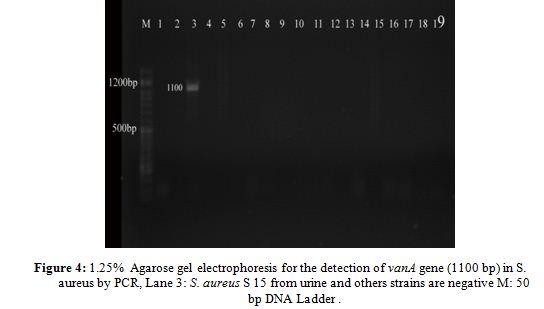
The sequenced vanA gene PCR product (Table 2, Fig. 4) was positive only in one strain of S. aureus S15 (VRSA) with percentage (2.5%) and gave a 1100 bp but none of the others. While for S. epidermidis it didn’t found vanA gene in any strain. In conclusion vancomycin resistance the almost strains of S. aureus were sensitive to vancomycin, so these results agree with genetic data where no strains of S. aureus carried vanA gene except S15.
These results like the results in some researches and unlike the others, where Kondoh et al. (25) showed that of 78 MRSA strains was one strain VRSA and five showed a hetero-VRSA pattern with 4–8 μg ml-1 for MIC; Adam et al. (26) detected and characterized the heterogeneous Vancomycin- Intermediate S. aureus were isolated in Canada and showed the differences between the value of MIC in these strains; Azimian et al. (27) isolated resistant strains of S. aureus from the respiratory tract of a patient in university hospital carried vanA with 713 bp and isolated some strains of CNS didn’t carry vanA gene; Entenza et al. (28) investigated the potential of microcalorimetry to detect reduced susceptibility to vancomycin in MRSA strains comparing with VSSA, VISA, VRSA as a reference clinical strains.
In conclusion, just the clinical S. aureus strains which have high value of MIC of methicillin or vancomycin, have mecA or vanA genes.
|
|
| References |
1-Kayser F. H. et al.
Medical Microbiology. Thieme Stuttgart, 10th Edition, 229-234, 2005.
2-Tortora G. J. et al.
Microbiology an introduction. Pearson, 11th Edition, 589-613, 2013.
3-Brooks F. G. et al.
Jawetz, Melnick, Adelberg’s medical microbiology, Lange McGraw-Hill, 24th Edition, 470-490, 2007.
4-Wolff K. et al.
Fitzpatrick’s color atlas and synopsis of clinical dermatology, McGraw-Hill, 6th Edition, 590-630, 2009.
5-Walker B. R. et al.
Davidson’s principles & practice of medicine. Churchill Livingstone Elsevier, 22th Edition, 329-349, 2014.
6-Palazzo I. C.V. et al.
First report of vancomycin-resistant Staphylococci isolated from healthy carriers in Brazil. J. Clin. Microbiol. 43(1): 179-185. 2005.
7-Wolk D. M. et al.
Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid xpert MRSA/SA skin and soft tissue and blood culture assays. J. Clin. Microbiol. 47 (3):
823-826, 2009.
8-Al-Obeid S. et al.
First detection of an invasive Staphylococcus aureus strain (D958) with reduced susceptibility to Glycopeptides in Saudi Arabia. J. Clin. Microbiol. 48 (6): 2199-2204, 2010.
9-Bhargava K.; and Zhang Y.
Multidrug-resistant coagulase-negative Staphylococci in food animals. J. Appl. Microbiol. 113: 1027-1036. 2012.
10-Gutierrez K. et al.
Staphylococcal infections in children, California USA 1985-2009. Emerg. Infect. Dis., 19 (1): 10-20. 2013.
11-De Matoze P. D. M. et al.
Antimicrobial synergism against different lineages of methicillin-resistant Staphylococcus aureus carrying of SCCmec IV. J. Appl. Microbiol.116: 1418-1426. 2014.
12-De vos P. et al.
Bergey’s manual of systematic bacteriology, the firmicutes. Springer, 2nd Edition, 19-124, 392-420, 2009.
13-Isenberg H. D. Clinical microbiology procedures handbook. ASM press, 2nd Edition, 127-150, 2007.
14-Benson.
Microbiological Applications laboratory manual in general microbiology. McGraw-Hills, 8th Edition, 257-261, 2001.
15-Japoni A. et al.
Modified DNA extraction for rapid PCR detection of methicillin-resistant Staphylococci. Iran. Biomedical J. 8(3): 161-165, 2004.
16-Kalia A. et al.
Method for extraction of high-quality and high-quantity genomic DNA generally applicable to pathogenic bacteria. Anal. Bioch. 275: 1-5, 1999.
17-Thompson J. M. et al.
Antibiotic resistant Staphylococcus aureus in hospital wastewaters and sewage treatment plants with special reference to methicillin-resistant Staphylococcus aureus (MRSA). J. Appl. Microbiol. 114: 44-54, 2012.
18-Thati V. et al.
Vancomycin resistance among methicillin resistant Staphylococcus aureus isolated from intensive care units of tertiary care hospitals in Hyderabad. Indian J. Med. Res, 134 (11): 704-708, 2011.
19-CLSI Clinical and Laboratory Standards Institute Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th Edition, Vol. 27 M2-M9, M7-A7, 2007.
20-Khan A et al.
Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J. Infect. Dev. Ctries. 1(3): 289-295. 2007.
21-Buzaid N. et al.
Methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary surgical and trauma hospital in Benghazi, Libya. J. Infect. Dev. Ctries. 5(10): 723-726. 2011.
22-Abd-Elhafez M. et al.
An outbreak of methicillin-resistant Staphylococcus epidermidis among neonates in a hospital in Saudi Arabia. J. Infect. Dev. Ctries. 5(10): 692-699. 2011.
23-Lyer A. et al.
High incidence rate of methicillin-resistant Staphylococcus aureus (MRSA) among healthcare workers in Saudi Arabia. J. Infect. Dev. Ctries. 8(3): 372-378. 2014.
24-Sergelidis D. et al.
Isolation of methicillin-resistant Staphylococcus spp. from ready-to-eat fish products. Lett. Appl. Microbiol. 59: 500 -506, 2014.
25-Kondoh K. et al.
Comparison of arbitrarily primed-polymerase chain reaction and pulse-field gel electrophoresis for characterizing methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 35: 62 -67, 2002.
26-Adam H. J. et al.
Detection and characterization of heterogeneous vancomycin-intermediate Staphylococcus aureus isolated in Canada: results from the Canadian nosocomial infection surveillance program, 1995-2006. Antimicrob. Agents Chemother. 54(2): 945-949, 2010.
27-Azimian A. et al..
Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a university hospital in Northeastern Iran. J. Clin. Microbiol. 50 (11): 3581-3585, 2012.
28-Entenza J. M. et al.
Rapid detection of Staphylococcus aureus strains with reduced susceptibility to vancomycin by isothermal microcalorimetry. J. Clin. Microbiol. 52 (1): 180-186, 2014
|
| |
| المجلد 8 ,
العددان 1 و 2
, رمضان 1437 - تموز (يوليو) 2016 |
|
|
|
